#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Steps_1_Correlation_Between_Samples~~~~~~~~~~~~~~~~~~~~~~~#
rm(list = ls())
workingDir = ".";
setwd(workingDir);
library(ggcorrplot)
library(openxlsx)
library(ComplexHeatmap)
library(ggplot2)
library(corrplot)
# loading data
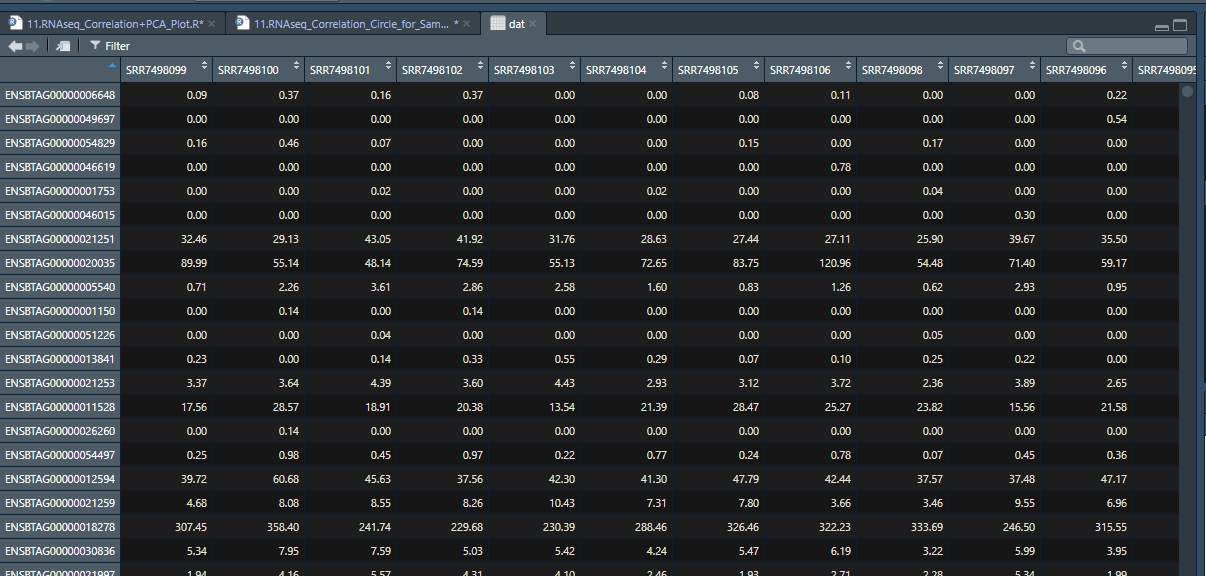
dat=read.csv("../Data/Angus.Rumen.Tpm.csv",row.names = 1)
head(dat,6)
names(dat)
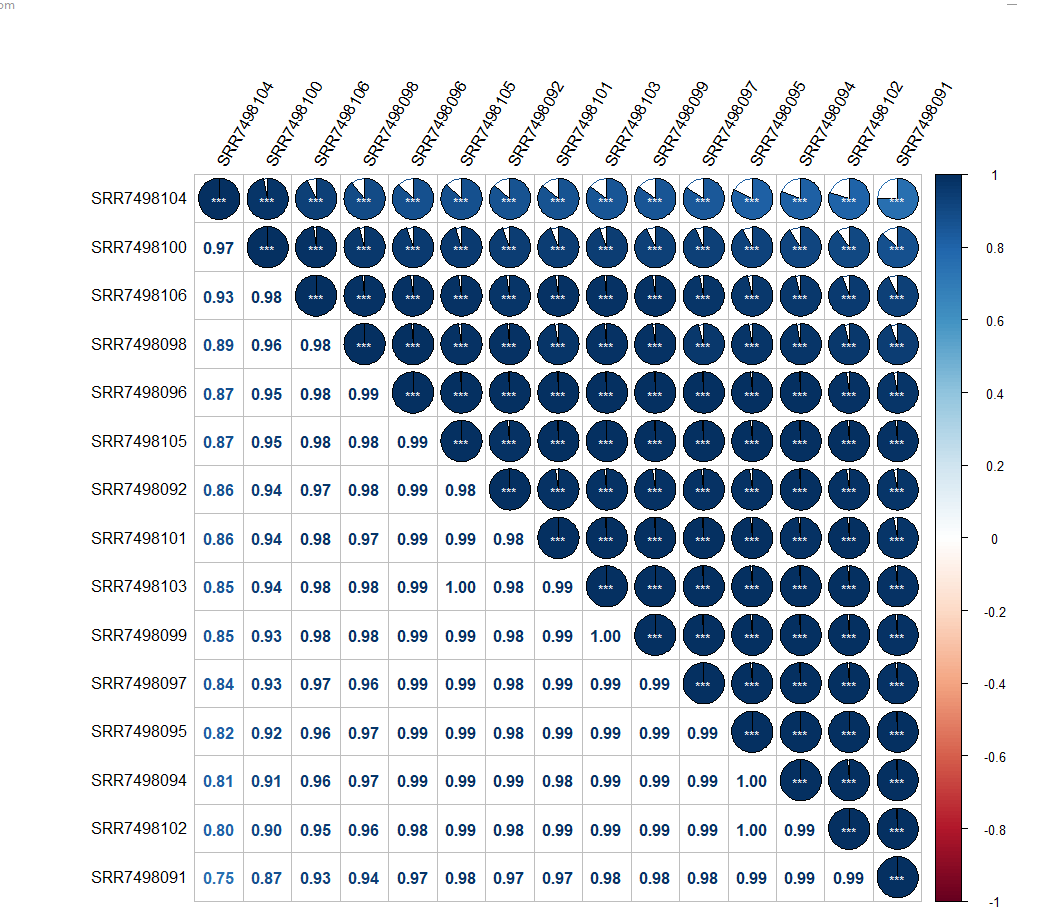
corr=round(cor(dat[,]),2) # Calculate the correlation coefficient
res1 <- cor.mtest(dat, conf.level = .95)
corrplot(corr, p.mat = res1$p, sig.level = .01)
## 设置不显著的空白
corrplot(corr, p.mat = res1$p, insig = "blank")
## 设置不显著的显示p值
corrplot(corr, p.mat = res1$p, insig = "p-value")
## 显示所有p值
corrplot(corr, p.mat = res1$p, insig = "p-value", sig.level = -1)
## 用 * 数量代表显示性
corrplot(corr, p.mat = res1$p, insig = "label_sig",
sig.level = c(.001, .01, .05), pch.cex = .9, pch.col = "black")
tiff(filename = "../Results/1.Rumen.cor.tiff",
width = 3,
height = 3,
units = 'in',
res = 600,
pointsize = 5)
corrplot(corr,
method = "pie",
order = "AOE",
hclust.method = "ward.D2",
addrect = 4,
rect.col = "blue",
p.mat = res1$p, insig = "label_sig",
sig.level = c(.001, .01, .05),
pch.cex = .9,
pch.col = "white",
tl.col = "black",
tl.cex = 1,tl.srt=60
)
corrplot(corr,add=TRUE, type="lower",
method="number",
order="AOE",
diag=FALSE,
tl.pos="n",
cl.pos="n")
dev.off()
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~#
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Steps_1_PCA_Between_Samples~~~~~~~~~~~~~~~~~~~~~~~#
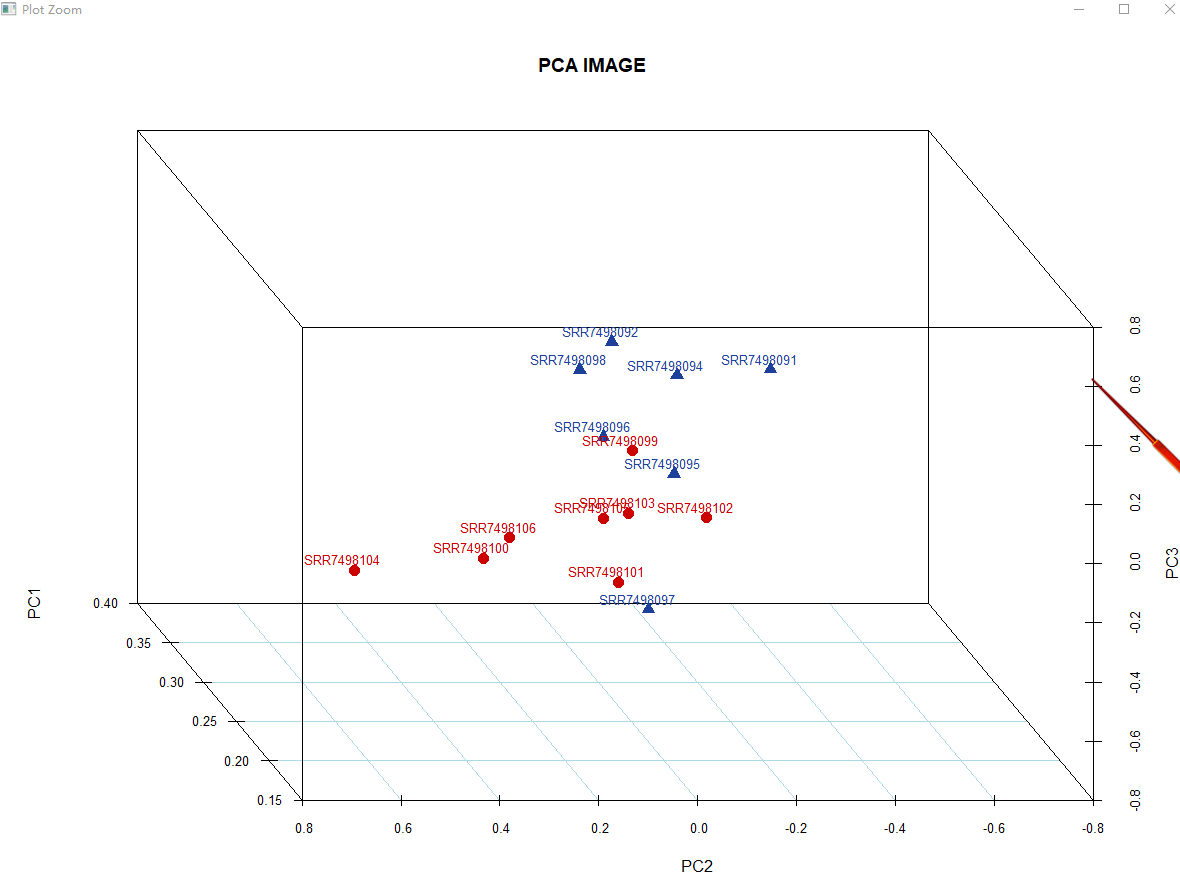
## PCA
library(gmodels)
pca= fast.prcomp(dat[,])
pca$rotation
## plots
colnames(dat)
library(scatterplot3d)
pca_info= data.frame(sample = row.names(pca$rotation),
Type=c(rep('LRFI',8),rep('HRFI',7)),
pca$rotation)
# colors and shapes
colors = c(rep('#CC0000',8),rep('#1d419b',7))
shapes = c(rep(16,8),rep(17,7))
###############################################
getwd()
tiff(filename = "../Results/1.Rumen.3D.pca.tiff",
width = 4,
height = 3,
units = 'in',
res = 600,
pointsize = 5)
s3d <- scatterplot3d(pca$rotation[,c(1:3)],
main='PCA IMAGE',
color=colors,
pch=shapes,
type='p',
highlight.3d=F,
angle=-60,
grid=T,
box=T,
scale.y=1,
cex.symbols=1.5,
col.grid='lightblue')
legend('bottom',legend = unique(pca_info$Type),
col = unique(colors),
pch = unique(shapes),
inset = -0.25,
xpd = TRUE,
horiz = TRUE)
text(s3d$xyz.convert(pca_info[,c('PC1','PC2','PC3')]+0.01),
labels = pca_info$sample,
cex = 0.8,
col = colors)
dev.off()
?pch
## plots from different angles
pca.plot <- function(ang){
scatterplot3d(pca$rotation[,c(1:3)],
main='PCA IMAGE',
color=colors,
type='p',
highlight.3d=F,
angle=ang,
grid=T,
box=T,
scale.y=1,
cex.symbols=1.5,
pch=shapes,
col.grid='lightblue')
legend('bottom',legend = unique(pca_info$Type),
col = unique(colors),
pch = unique(shapes),
inset = -0.25,
xpd = TRUE,
horiz = TRUE)
text(s3d$xyz.convert(pca_info[,c('PC1','PC2','PC3')]+0.01),
labels = pca_info$sample,
cex = 0.8,
col = colors)
}
pdf("../Results//1.Rumen.PCA.pdf",height = 6,width = 6)
sapply(seq(-360,360,5),
pca.plot)
dev.off()数据格式: